CLPP(ID:7996/)
Cellular list procssing language
for Cellular List-Processing Program.
Pioneering Bioinformatic system developed by Stahl et al at Oregon State 1963-1969, actually given a formal name around 1965. It was the source language for the TASP (Turing Automaton Simulation Program) and ran on the SDS-920 computer.
It had pattern matching capabilities in 2-dimensional structures as well as references to morphogenesis (after Turing's celebrated paper). Involved the creation of organic algorithms to represent (eg) genetic adaptation, so possibly the first use of such tools in computing. (Certainly it is the first language to feature these as a top level concept).
List processing lanugage with special facilities for cell simulation, based on Post and Turing systems, with extra features from COMIT and IPL.
Related languages
| COMIT II | => | CLPP | Evolution of aspects | |
| IPL-IV | => | CLPP | Evolution of aspects | |
| Post production | => | CLPP | Influence | |
| Turing's code | => | CLPP | Influence |
References:
in [AFIPS JCC 24] Proceedings of the 1963 Fall Joint Computer Conference FJCC 1963, Las Vegas, Nev., Nov. 1963, view details
in [AFIPS JCC 24] Proceedings of the 1963 Fall Joint Computer Conference FJCC 1963, Las Vegas, Nev., Nov. 1963, view details
in [AFIPS JCC 24] Proceedings of the 1963 Fall Joint Computer Conference FJCC 1963, Las Vegas, Nev., Nov. 1963, view details
Introduction
In the last few years enormous progress has been made in clarifying the operational mechanisms of living cells. It has been established beyond reasonable doubt that all aspects of cell activity are controlled by sequences of elementary genetic units. A comma-free triplet coding in the four-letter alphabet of DNA is transcribed on RNA and causes the formation of sequences of amino acids, which make up polypeptides and proteins. Various theories of transcription control for such systems are now under study. Recently, synthetic nucleic acid (RNA) chains have been fed into the cell machinery, thus demonstrating that protein synthesis can be controlled artificially. Numerous finer details of the problem could be mentioned [...] but shall not be considered in this report.
There arises the question of what type of mathematical modelling method is best suited for simulation of molecular genetics. In the past numerical models, based on chemical kinetics expressed in terms of simultaneous differential equations, have usually been applied.
Impressive results were obtained by Chance et a1., Garfinkel, Hommes and Zilliken and others. However, it has also become clear that simultaneous solution of hundreds or thousands of differential equations, many of whose coefficients probably cannot be measured experimentally[...], poses a difficult problem. Actual cells may contain thousands of different genes and hundreds of thousands of synthetic units. Major questions of solvability and stability of such systems must be faced in an attempt to model a complete cell.
The present report describes a fundamentally different approach to the problem of simulating a cell, some aspects of which were reported earlier in Stahl and Gheen. Since genes and proteins are representable as linear chains or strings, it is proposed that molecular mechanisms of cells be simulated by string-processing finite automata. In this model strings representing DNA, RNA, proteins and general bio-chemicals are subjected to controlled copying, synthesis into longer strings and breakdown into shorter ones, with use of what may be called "algorithmic enzymes." A major property of these logical operators is that they are combinable into large systems with complex properties.
The materials below deal in turn with a new computer simulation system for studying finite automata, the properties of algorithmic enzymes, experimental studies with systems of the latter and lastly with some questions of solvability for model cells defined entirely by automation-like enzyme operators. Extract: The Automaton Simulation System
The Automaton Simulation System
The quintuplet command code proposed by Turing is used in the programming system and well described in Turing Davis and elsewhere. Turing's device was designed for proofs of computability and in principle requires an infinite tape and infinite number of recursive steps for such demonstrations. This circumstance does not prevent one from using the quintuplet code for general purpose simulation of automata. The late John von Neumann pointed out that the Turing Machine represents a means of programming or simulating any algorithm, as well as for computability demonstrations. McNaughton has emphasized that the Turing Machine should be considered as the most general of automata.
A compiler based on the Turing quintuplet notation (but not really modelling the Turing Machine) has been designed and is described in Coffin, Goheen and Stahl. Simulation of a considerable number of different automata on the system, including ones for pattern recognition, has revealed that a computer program for modelling of automata is a useful research tool, which may find practical applications when it is desired to use a "variable programmed automaton."
Results substantiating this conclusion are reported in Stahl, Coffin and Goheen and Stahl, Waters and Coffin. A special compiler, constituting an "automaton simulation program," was written for a SDS-920 computer and has processing rates of up to 10,000 quintuplet commands per second. Rates of over one million automation commands a second should be possible with presently known technology. A number of special provisions have been included for automatic sequencing of different circumscribed algorithms or automata presented as lists of quintuplets, debugging, selective printout of string during simulation runs, and so forth.
A Turing compiler should not be evaluated on the basis of the inefficient operation of most Turing Machines described in the literature to date. The authors are using individual Turing Programs (algorithms) exceeding 1700 quintuplets in size and a complete system (namely, the algorithmic cell), which includes over 43,000 quintuplets. Interesting results have been obtained for the problem of recognition of hand-printed letters (A-2) and shall be reported elsewhere (Stahl, Waters and Coffin).
Naturally, automaton simulation has a special range of application, as do research compilers such as LISP, IPL-V, or COMIT.
The Turing Machine is a device which operates on individual symbols presented on a single long tape, along which a reading head moves left or right, one cell square at a time. A capability is provided for erasing and writing individual symbols and for recording the "state" of the Turing Machine, which defines uniquely its response to a particular viewed symbol.
Only one type of program command is used and consists of a quintuplet (or matrix table with output triplets of symbols), which usually appears as follows: symbol scanned, state of machine, new state, motion (right-R, left-L and stay in place-P) and symbol to replace existing symbol before motion is carried out. A quintuplet such as 12 A:15RB is read "if in state 12 and A is viewed, then replace A by B, go to state 15 and move right one square." A final logical halt of the automaton takes place on such an entry as 26* :26P*, which is read "if asterisk is seen in state 26, remain in place, stay in state 26 and do not alter asterisk." In principle, the Turing Machine must have available an infinite tape and amount of time, but precisely the same notation can be used with finite automata and this has been done by such authors as McNaughton, Myhill, Trakhtenbrot and others. The quintuplet command structure need not in itself connote the extended and often inefficient "shuffling" operation of the classical Turing Machine. Extract: Algorithmic Enzymes
Algorithmic Enzymes
The concept of a Turing quintuplet code may be illustrated with a simple but biologically provocative example in which a finite automaton simulates a lytic enzyme that breaks down strings. Table I is a quintuplet program for an "automaton enzyme" which lyses strings in the alphabet (AGCT), representing the four nucleic acid bases adenine, guanine, cytosine and thymine. A typical input tape into the automaton using this code might be
... Ø Ø = A – C – G – C – C – T – T – A – G – C – A = Ø Ø ... (1)
in which "Ø"-empty cells, "="-start of string, "–"-bond between letters.
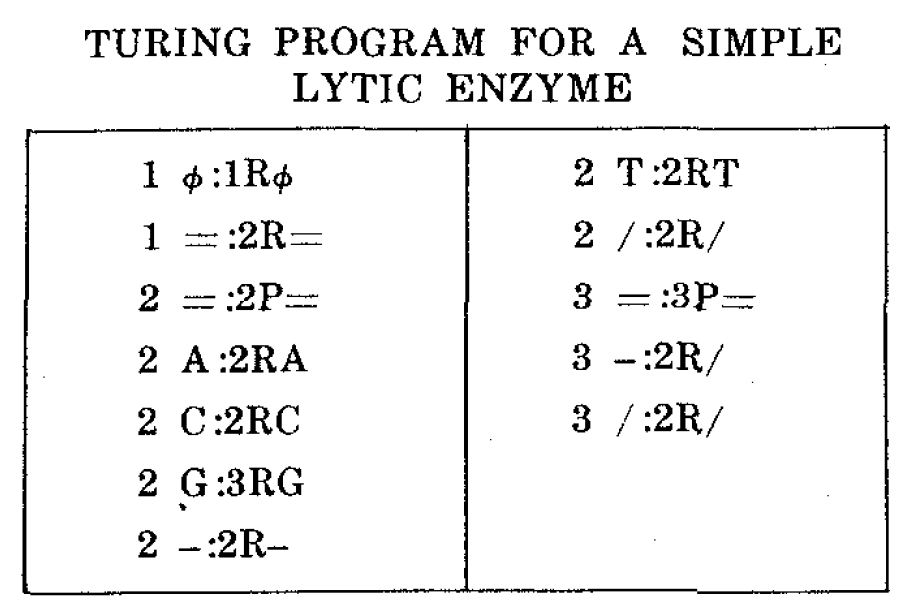
Following a single left to right pass the string in (1) will be converted into
=A-C-G/C-C-T-T-A-G/C-A= (2)
in which every bond immediately to the right of a G, regardless of what symbol is next to it on the left or right, is converted into an "open bond" (/).
Operation commences at the left end of the string. The empty symbols (Ø) are passed over by entry 1Ø:1RØ. When the left end-marker (=) is seen control passes to state 2. In state 2 all symbols except G (namely, A,C,T and -) are simply skipped over, as in 2-:2-. If G is seen, by entry 2 G:3RG, control passes to state 3, which next encounters a "bond" and converts it to an "open bond" using entry 3-:2R/. Provisions are also made for skipping over any existing open bonds, as in entry 2/:2R/. Stopping occurs in state 2 or 3, on an entry such as 3 =:3P=.
Table II is a sample of coding for a string synthesizing finite automaton, which was described in Stahl and Goheen and is the prototype for the algorithmic enzymes noted in this paper. The cited work also includes automata for copying and complementary copying of strings (as in DNA transcription). and for more complex types of lysis.
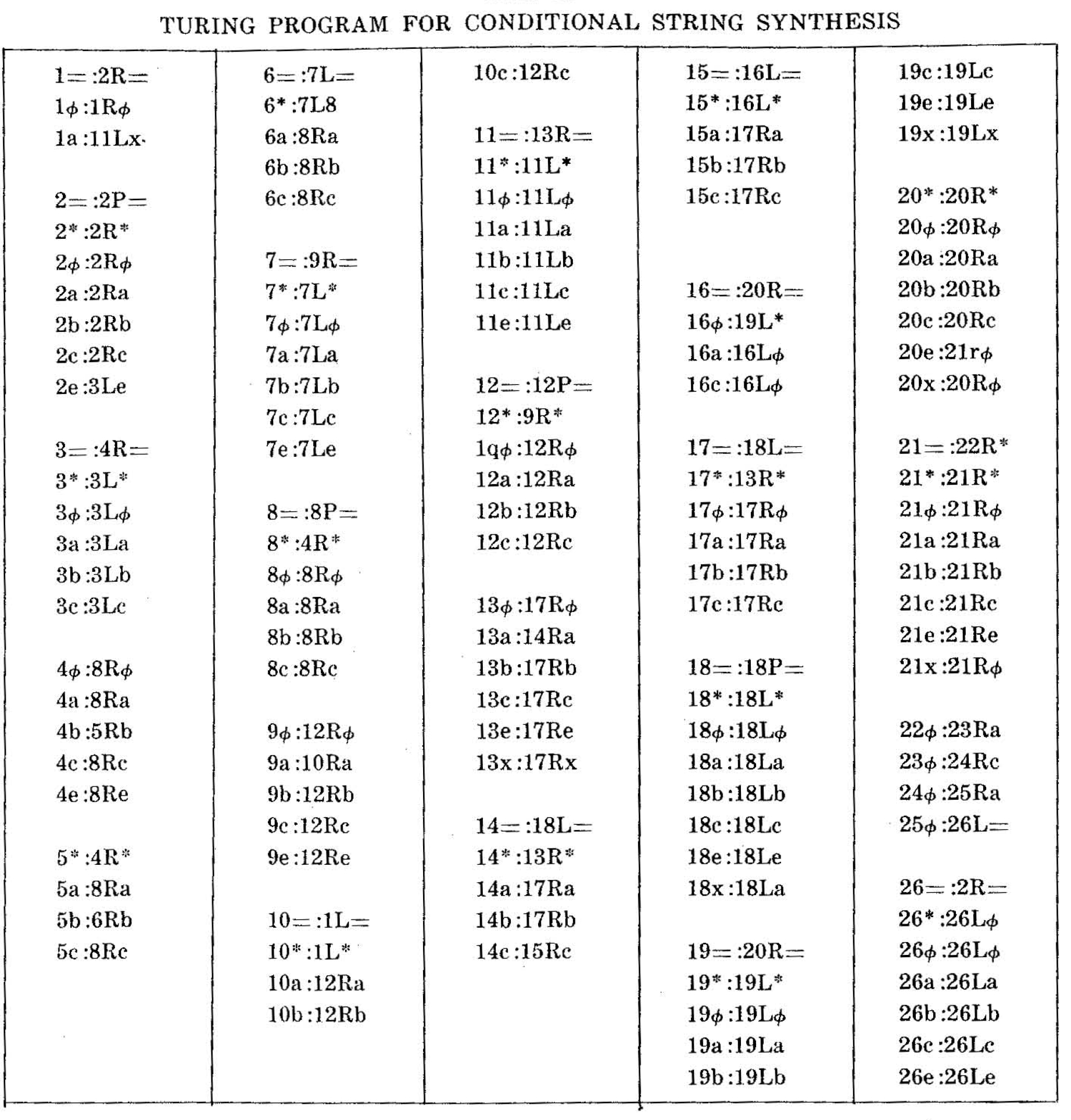
In general, the construction of quintuplet programs for automata is straightforward. It is noteworthy that they are truly interchangeable because of the very standard format. It is clear, however, that string-processing enzymes might also be represented by other types of automata, such as Wang's68 modified Turing Machine and that it would be entirely feasible to design special compilers that accept a "statesymbol" table.
While the lytic enzyme of Table I was given principally as an example, it is interesting to note that reconstruction of a parent protein string following the action of several lytic enzymes is an important problem today for nucleic acid and protein analysis. Rice and have pointed out that application of three specific lytic enzymes, which split "next to" only three of the four specific bases in DNA, does not allow a unique reconstruction of the parent chain. This is an excellent example of algorithmic unsolvability arising in a biological context, and moreover even in very classical form, namely, solution of a "word problem" by algorithmic methods.
It must be emphasized that the lytic enzyme of Table I in no way models the physiochemical properties of any real enzyme that might perform the indicated lysis, and only simulates the string-processing aspects of the enzyme action. This type of model is somewhat comparable to the McCulloch-Pitts imitation of neurons by the threshold Boolean "logical neuron," in that both model systems are rather gross from the biological viewpoint, but involve a consistent mathematical methodology. The McCulloch- Pitts neuron can be combined into large systems, such as perceptrons, and much the same $ten has been taken with algorithmic enzymes. The main problem in biological modelling is probably that of finding well-defined mathematical methods which can be applied with profit to the biological system.
The concept of studying cells algorithmically was probably implied in von Neumann's work on self-reproduction of structures composed of finite automata. This model has been extensively analyzed and extended by Burks and Moore. The growth and stability of automaton-like arrays is considered in Lofgren, Ulam, Eden, Blum and others. Rashevsky suggested the application of the Marltov "Normal Algorithm" (A. A. MarkovZa) to the genetic codes, but did not define any complete cell model. Pattee's proposed that a simple automaton could produce long biological chains of a repetitive kind. Induction and repression mechanisms in cells are analyzed by Jacob and Monod. Sugita and Rosen explore the Boolean logical representation of cells. Turing himself wrote a paper entitled "The Chemical Basis of Morphogenesis," but used differential equations rather than algorithm theory in this study. Soviet workers such as Frank1 and Pasynskiy discuss some general aspects of cellular control theory. Lyapunov proposes the systematic study of interacting automata subjected to executive hierarchies of control and Medvedev deals with errors in genetic coding.
An "algorithmic cell" is defined in this report as a system of string-processing finite automata, representing enzymes, together with the cell contents, identified as strings. Smaller biochemicals which are not as obviously strings as DNA or proteins may be coded as strings. This is done routinely in ordinary chemical nomenclature.
Furthermore, the linearly-coded enzymes (proteins) must be able to recognize any normal biochemical in the cell, and this may be interpreted to mean that some sort of a string coding of biochemicals is possible. Biochemists represent DNA, RNA and proteins in an associative symbolic notation, but this fact has not been subjected to mathematical interpretation. That is, molecular biologists now assume that the four DNA bases, the four RNA bases and the 20 or so pertinent amino acids can and should be presented as symbols in an associntive (parentheses free) linear string notation. Much progress has been made recently in showing the details of the nucleic acid to protein coding (NirenbergJ1). It is noteworthy that a normal algorithm is implied in the DNA triplet to amino acid substitution coding, e.g., CAC 'Pro. (cytosine-adenine-cytosine codes the+ amino acid proline). Although the DNA to protein coding is understood, almost nothing is known about the grammer, syntactical relationships or programming techniques used by the cell during its strictly deterministic, and therefore algorithmic, growth, differentiation and functioning.
Figure 1.1 is a flow chart of string syntheses taking place in a typical cell model of the type suggested in this report. At present 43 separate threshold algorithmic enzymes are used and produce a total of nine different final products.
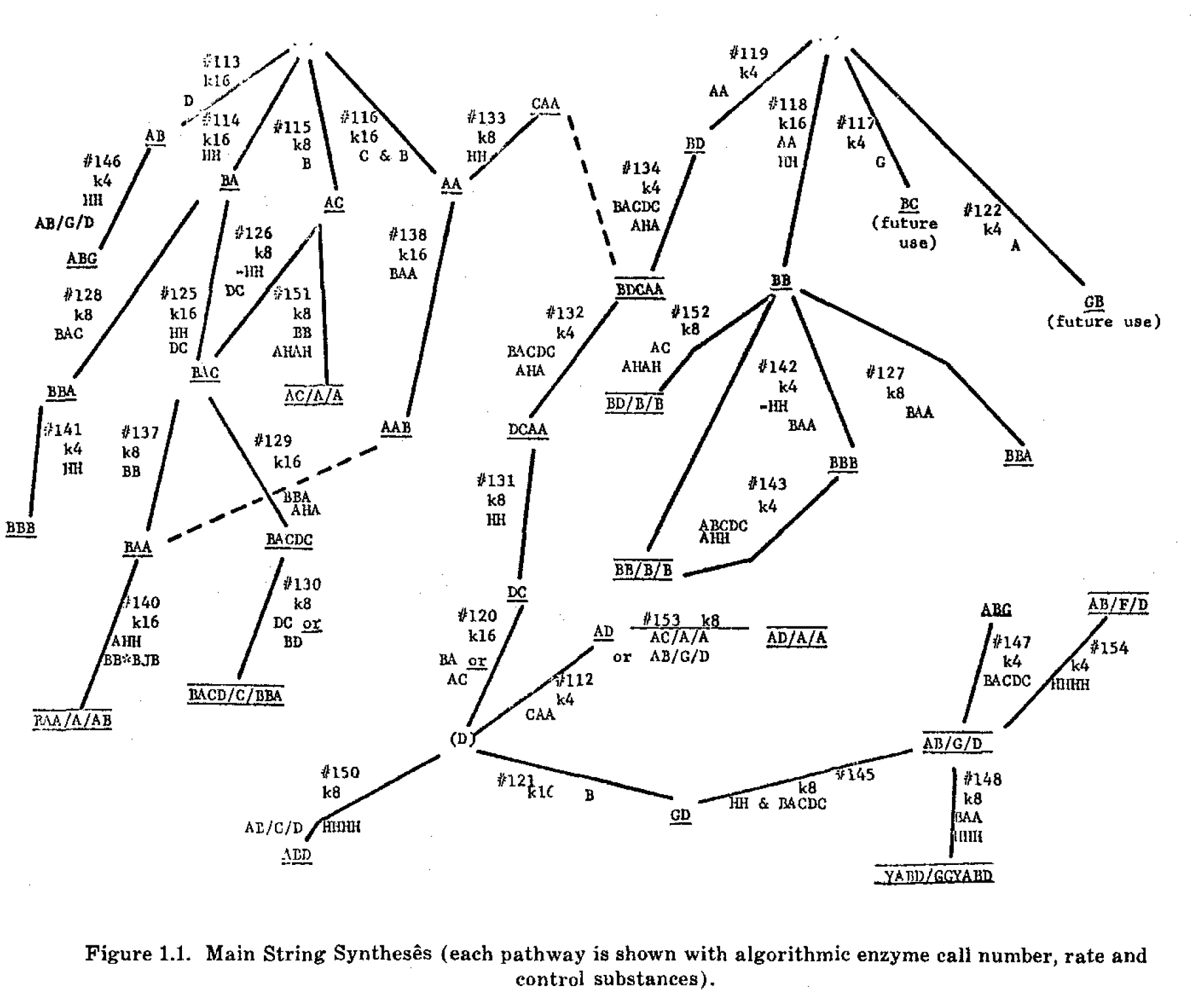
The complete model cell consists of the enzyme operators stored on magnetic tape, a 2000 symbol memory which has a place for each string and its numbers at any moment of operation, and three "housekeeping programs" (written entirely in quintuplets representing specialized finite automata) which perform addition, generation of pseudo-random variables, totalling operations, counting, etc. While this type of programming is obviously not as efficient as a conventional computer methods for straightforward arithmetic operations, it is a consistent automaton simulation methodology and very flexible. For example, total cell size may be controlled by simply introducing a special automaton that changes rates of "diet" letter entry as a function of total cell string count. A complete growth experiment with an algorithmic cell requires about 30-45 minutes and involves hundreds of thousands of individual enzyme steps.
The algorithmic enzyme used in the cell model is more complex than the one defined by Table I1 and functions as follows. Each enzyme is stored as a program of about 1000 quintuplets on magnetic tape and called into the core memory by the compiler program when a preceding enzyme or a "housekeeping automaton" writes in a "call number" in a specific region of the memory tape. As the first step, threshold checks are made of "energy" represented as a simple string and of one or two control strings, by interrogation of the binary numbers associated with these strings. If these thresholds are met it is next ascertained that the substrates (input materials) for the specific enzyme are available in sufficiently high quantities and that the final product is not already present in excessively large amounts. If these added threshold conditions are met, the enzyme functions, producing a fixed quantity of its product, while removing appropriate quantities of the materials that went into the latter. After this is done the algorithmic enzyme writes in another "call number" which controls which housekeeping program or other enzyme shall function next.
In Figure 1 the enzymes are identified by number, as #113, and the "k" value indicates synthetic rate for a unit cycle, while the strings along the flow lines are the control substances.
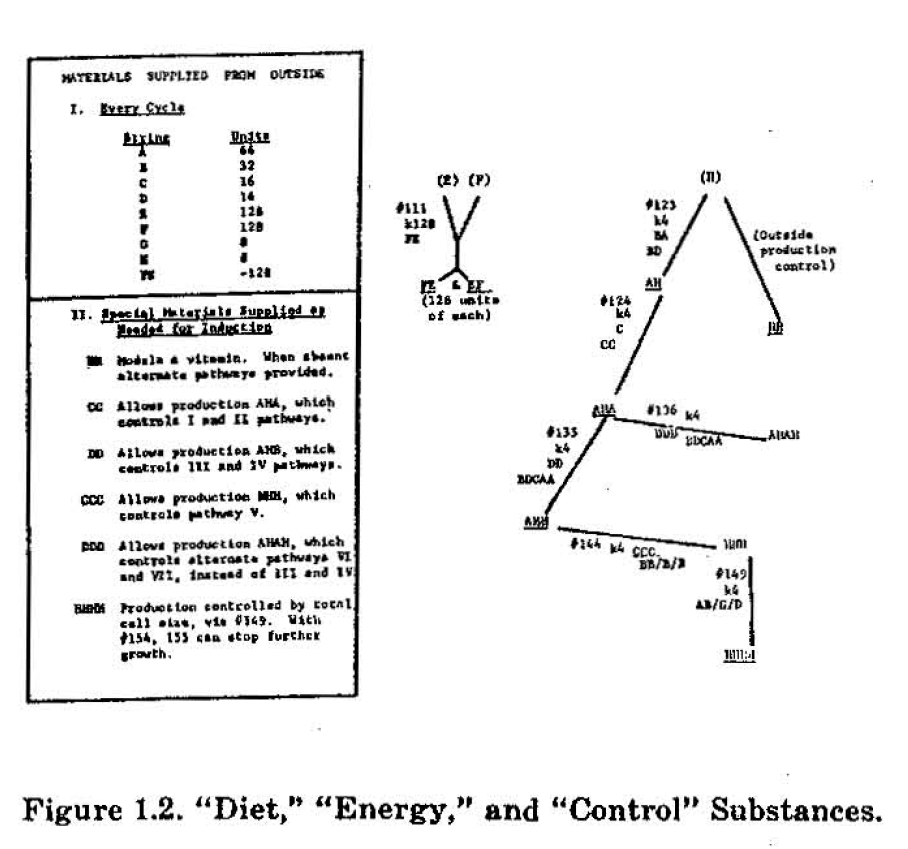
A complete set of specifications for the enzymes shown in Figure 1 will be published elsewhere. It can be seen from Figure 1 that certain "inducer strings," such as AHA, HHH, etc., control the action of sets of coordinated reactions. Moreover, synthesis of these "control strings" is subjected to induction or repression as a function of certain special strings that may be supplied from the outside, as by an adjacent model cell, or manufactured in the cell if levels of "metabolite strings" meet certain arbitrary thresholds. A reasonable correspondence can be drawn between the activities of the algorithmic cell model and known basic cell activities, including blocking and unblocking of DNA, formation of messenger RNA, synthesis of enzymes and proteins on ribosomes, production of metabolites by enzymes, etc.
The flow chart of Figure 1 is entirely arbitrary and does not attempt to represent any actual cell. It must be noted that a real cell may have tens of thousands of enzymes and genes, and that firm quantitative data is available at 'present only for limited sets of enzymatic reactions. Nevertheless, one may study the basic mathematical problems presented by string-synthesizing automata. The final products of the algorithmic cell model, as shown in Figure 1 were designed to combine into several different two-dimensional arrays. An example of a unit in such an array is given in Figure 2. A "complementation algorithm" is applied to the product strings. This states that any two strings will stick together or polymerize providing that they have at least three complementary bonds, such as A with B, B with A, C with D, etc. Presence of a non-complementary bond negates two complementary bonds. This may be considered to be still another kind of discrete threshold action that generates complex entities from simplier units.
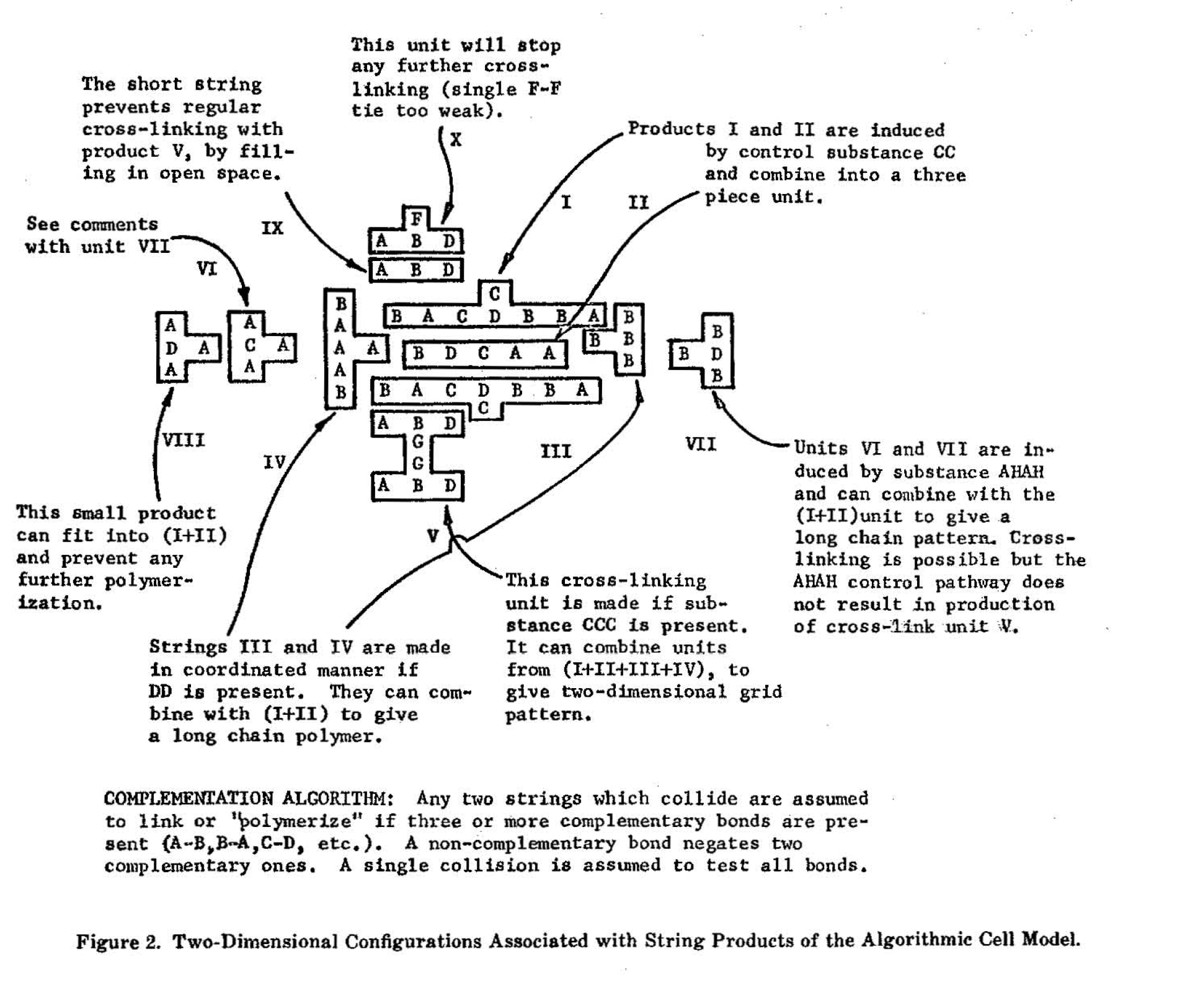
The control strings are so chosen that they regulate the coordinated formation of one or more final products that polymerize together. There are sufficient product strings so that different types of arrays are possible and may lead to long individual chains, cross-linked chains, circumscribed blocks of strings and even grids with a special border that limits further growth. All the operations leading to these results are defined by firm algorithms. However, pseudorandom variables are added to generation rates and this results in less "mechanical" action.
in [AFIPS JCC 25] Proceedings of the 1964 Spring Joint Computer Conference SJCC 1964 view details
Introduction (part 1)
Recent breakthroughs in molecular biology indicate that living cells are very complex chemical machines whose activities are programmed by their genes. When faced with some environmental hazard a cell might attempt to design a new gene. The problem can be studied by using a cell model based on automata theory and simulated on a computer. The model suggests that a molecular adaptive automaton within a cell required to compute the DNA coding for a new gene would be faced by inherent logical paradoxes or algorithmically unsolvable problems. Real cells are not believed ever to accomplish direct genetic adaptation and evolution proceeds only through Darwinian natural selection.
Previous reports described details of cell models processed on a special compiler for a SDS-920 (Turing Automaton Simulation Program - TASP) and designed for the study of systems of finite automata. Individual automata of over 1,000 standard (Turing quintuplet) commands are used, with systems of 40-100 such automata linked together by automatic procedures. The automata move freely about a memory tape which may be up to 2,000 symbols in length, at operating rates of 1,000-10,000 commands per second. The TASP system has convenient provisions for editing, printing, etc., and has been in steady operation since 1963.
The system was first used to investigate a cell model based on 43 "enzyme automata," which demonstrated biological phenomena such as demonstrated biological phenomena such as growth, differentiation, and resistance against enia vironmental instabilities and injury. It is a useful working example of a self-organizing system and interesting from the viewpoint of biological cybernetics. Recently another type of model was completed which demonstrates self-reproductIon in the sense proposed by von Neumann and subsequently extended by Moore and others. This system consists of 35 gene-enzyme automata acting upon some 46 different "biochemical strings," represented on a long cell contents tape. Reproduction of the linear cell simulates a number of processes observed in real cells and requires about 80 minutes of computing time on the SDS-920.
Extract: Basic operation
The basic logical operator of the cell model is what might be described as a fixed-rate, multiple-threshold, string-recognizing, string-synthesizing completely standard (axiomatic) manner, using Turing Automaton notation. The TASP simulator functions as a Universal Turing Machine, into which is fed the linear representation of the total cell contents together with the gene coding. The cell automata are able to move freely about the cell contents string, including, in principle, the actual coding regions (genes) for all automata stored in memory. To restrict length of the cell tape the genes are actually represented by code numbers which serve to identify and also call into core memory the Turing codes for each gene, but these have the same operational and symbolic significance as the lengthy Turing Automata numerical sequences.
Extract: Modelling genetic adaptive behaviour
In order to study simulated genetic adaptation, the self-reproducing system design includes a special gene which represents a molecular adaptive automaton" (there is no evidence for such a device in real cells). This functions in exactly the same manner as all the other cell automata and is activated by evidence of potential cell injury, such as low energy, absence of elementary diet units, missing "wall symbol" etc., which might result from starvation, coldness, or attack by a virus. This model and its biological interpretations have been found acceptable by molecular biologists.
The hypothetical adaptive automaton needs first to be constructed from subunits (strings) within the cell, in the same manner as any other enzyme automaton, but this may often be impossible, e.g., during starvation. Assuming, however, the test automaton has been produced it must scan the cell contents string, and the coding of all cell automata (including itself) and then compute the coding for a new adaptive gene-enzyme. This task can be stated specifically in terms of choosing the n + 1 amino acid symbol or n + 1 DNA nucleotide symbol for a partially completed enzyme or gene of n units. In the limiting case the test automaton may be assumed to have the power of a Turing Machine rather than a standard string-processing finite automaton.
When formulated in this abstract manner the problem of direct genetic adaptation is seen to require solution of classical unsolvable problems, such as the Halting, Applicability, Self-Applicability and Minimality Problems of the Turing Machine. For example the cell test automaton must predict if the tentative gene string for a new enzyme automaton is fully compatible with normal operation of the existing system of automata under all possible circumstances. To give a biological example, one would have to deduce from the DNA sequence for a new proteolytic enzyme, designed perhaps to attack a virus, if it will break up any normal protein of the cell, including those in the test automaton.
Genetic unsolvability might also be approached by other classical methods, such as implicit in the Entscheidung's problem, Godel's proof, Richard's Paradox, and prediction problem in productive string systems, e.g., Post and Markov algorithms. From this viewpoint, the genes may be regarded as a system of "biochemical axioms" and it is necessary to prove consistency (survival or self-reproductive value) in a new augmented system of cell logic containing an added gene. Previously unformalized unsolvable problems appear to be involved in predicting whether a system of automata has a certain new property, and particularly if it is capable of self-reproduction. Cancer is believed to represent a failure of reproductive control algorithms and identification of a malignant cell appears to involve unsolvable identification problems for the cellular test automaton.
There is really no way "to prove" that the described model is a valid representation of a living cell, so the genetic unsolvability concept must be regarded as a hypothesis. Real cells are enormously more complex than any cell models available at this time, but are not thought ever to design new genes. Some portion of the genetic adatation problem appears be solvable, but evidently the entire procedure is not "computable" for any molecular automata. Mechanization of theorem proving has been shown by Wang (1963) to be a powerful tool; this paper suggests mechanization of cellular genetic adaptation and control problems. In any event, it is clear that molecular genetics is an important area of study for modern automata theory.
in Kalenich W.A. (ed), Proceedings IFIP Congress (New York, May 24-29, 1965), Spartan Books, New York, 1965. view details
in Kalenich W.A. (ed), Proceedings IFIP Congress (New York, May 24-29, 1965), Spartan Books, New York, 1965. view details
in Kalenich W.A. (ed), Proceedings IFIP Congress (New York, May 24-29, 1965), Spartan Books, New York, 1965. view details
in Kalenich W.A. (ed), Proceedings IFIP Congress (New York, May 24-29, 1965), Spartan Books, New York, 1965. view details